AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling
![Tutorial] How to perform docking of zinc metalloproteins using Autodock Vina? — Bioinformatics Review Tutorial] How to perform docking of zinc metalloproteins using Autodock Vina? — Bioinformatics Review](https://bioinformaticsreview.com/wp-content/uploads/2021/10/zn.jpg)
Tutorial] How to perform docking of zinc metalloproteins using Autodock Vina? — Bioinformatics Review
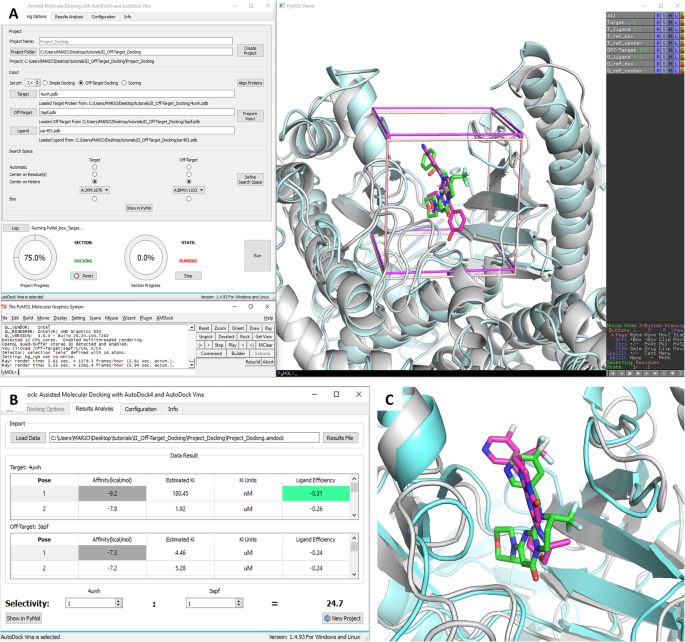
AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4 | Biology Direct | Full Text

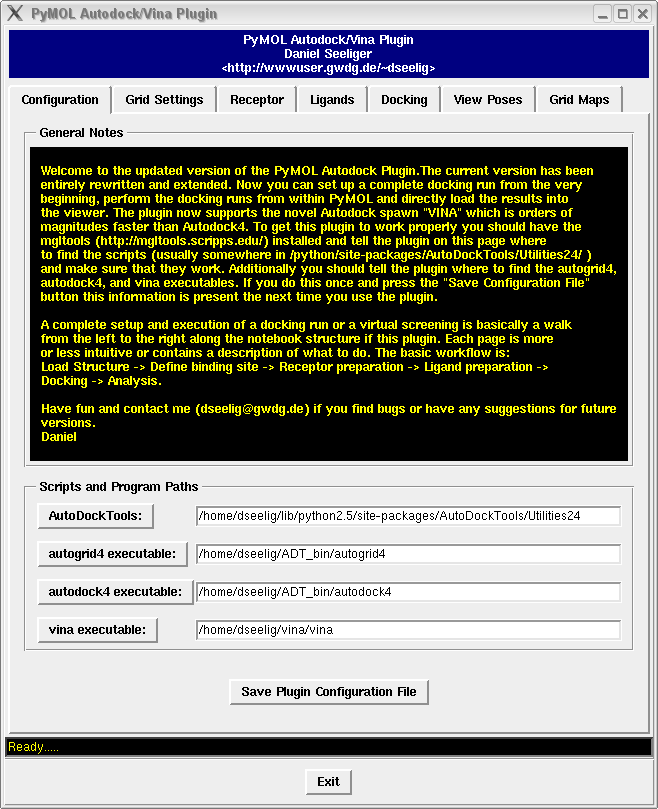
Autodock tutorial VINA with UCSF Chimera José R. Valverde CNB/CSIC © José R. Valverde, 2014 CC-BY-NC-SA. - ppt download